Authors: Marissa E Powers PhD, Jer-Ming Chia PhD, Priyanka Sebastian, Keith Mannthey
Background
The Genomics Analytics Toolkit (GATK), developed and maintained by the Broad Institute of MIT and Harvard, is a commonly used set of tools for processing genomics data. Along with the toolkit the Broad Institute publishes Best Practices pipelines, a collection of workflows for different high-throughput sequencing data analyses that serves as reference implementations.
To understand how the choice of Virtual Machine (VM) families affect the cost and performance profiles of GATK pipelines, the Solutions team at Intel, supported by the Azure High-Performance Computing team and Biomedical Platform team in Microsoft Health Futures, profiled a GATK pipeline on different Intel-based VM families on Azure. Based on this work, users can reduce cost by a 30% from $13 to less than $9 per sample simply by setting the workflow to run exclusively on VMs with later generation Intel processors. The Best Practices Pipeline for Germline Variant Calling performs alignment of fragments of DNA for a given sample, and then identifies specific mutations or variants in that sample. This pipeline is commonly used in many diagnostics and research laboratories and was selected as the focus for this benchmarking exercise. For more details on this pipeline, see here.
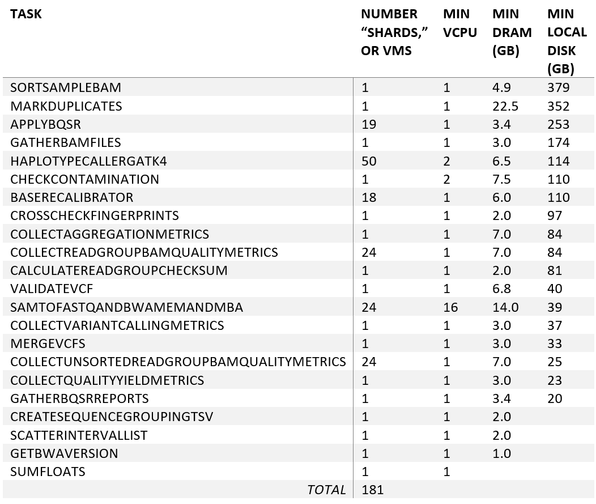
The pipeline consists of 24 tasks, each with distinct compute requirements. Of these 24 tasks, six get distributed as multiple jobs or, on Azure, on multiple VMs. As an example, SortSampleBam is executed on a single VM, while ApplyBQSR is distributed across 19 VMs.
Table 1 below provides a summary of the tasks in the pipeline, the number of VMs each task is distributed across, and the minimum number of vCPUs, DRAM, and disk required per VM for that task. Summing up all the VMs across all the tasks, there are a total of 181 VMs automatically orchestrated throughout the execution of the pipeline.
Optimal cost efficiency for this workload on Azure requires (1) efficient orchestration of these tasks; (2) ensuring each task is allocated the right-size VM; and (3) choosing the best VM series.
Table 1
GATK Pipelines on Azure
To enable efficient distribution of Best Practices Pipelines both on local infrastructure and on the cloud, the Broad Institute developed Cromwell, a workflow management system that can be integrated with various execution backends. Microsoft built and maintains Cromwell on Azure, an open-source project that configures all Azure resources needed to run Cromwell workflows on Microsoft Azure, and implements the GA4GH TES backend for orchestrating tasks that run in Azure Batch. Microsoft also maintains common workflows for running the GATK Best Practice Pipelines on Azure.
Figure 1, Ref: https://github.com/microsoft/Cromwell on Azure
Three different configurations for the Germline Variant Calling pipeline, each using different sets of VM series, were benchmarked to determine the most performant and the most cost-effective VM series for this workload.
In the “Default Configuration”, the least expensive VM in terms of $/hour was selected at runtime for each task. As an example, for the HaplotypeCaller task, the least expensive VM series that meets the 2 vCPU, 6.5GB DRAM, and 114GB disk requirements is the Standard-A VM series.
For “Dv2,” the smallest Dv2 VM that met the compute needs for each task was chosen. Likewise, for “Ddv4,” the smallest Ddv4 VM for each task that met the compute needs for the task was chosen. The Intel CPU generation for each of the configurations is shown in Table 2.
Table 2
Germline Variant Calling: Price/Performance on Azure
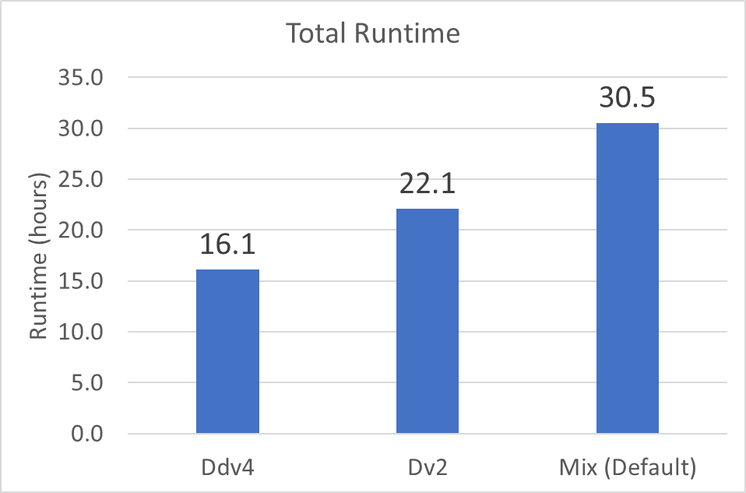
Figure 2 shows the end-to-end runtime for the GATK Best Practices Pipelines for Germline Variant Calling on each of the three configurations tested . The Ddv4 configuration had the fastest runtime at 16.1 hours . The Default configuration, where the least expensive VM is chosen for each task at runtime, results in a 1.9X slower runtime compared to VMs with the newer generation Intel processors . The Dv2 configuration, ran on VMs with newer generation CPUs compared to the Default configuration, results in a runtime of 22.1 hours.
Figure 2
While the germline variant calling pipeline performed better on Azure VMs with newer generation Intel CPUs, the cost of running the pipeline may hold higher priority for users without time-sensitive workloads. The Default configuration chooses the VM with the lowest in $/hour specifically for this reason.
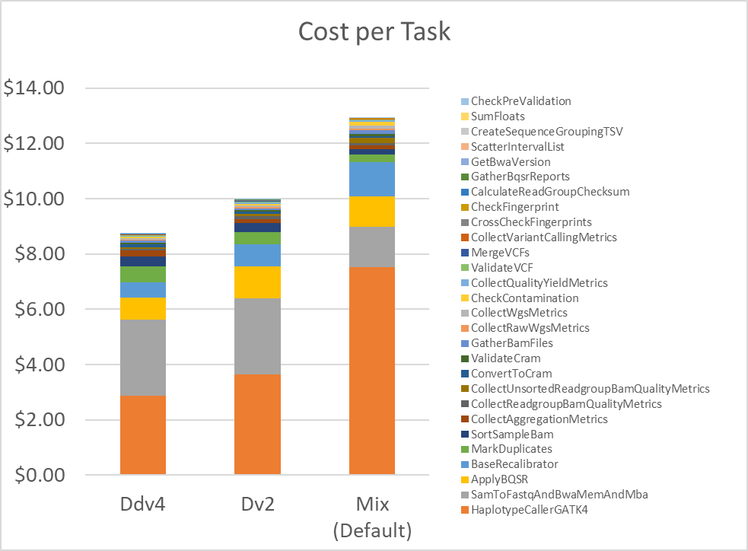
Figure 3 shows the cost to run the entire pipeline on each of the three configurations tested, broken out by cost per individual task . Perhaps counter-intuitively, the Ddv4 configuration with the highest $/hour VM, is the least expensive option. Users can reduce cost by a 30% from $13 to less than $9 simply by setting the workflow to run exclusively on the Ddv4 VM series instead of choosing the lowest $/hour VM.
Figure 3
The biggest cost savings is with HaplotypeCaller . With the Default configuration, HaplotypeCaller runs on Standard-A machines. As shown in Figure 4, this results in a cost of $7.53 specifically for this one task. When running with Ddv4, this cost is reduced to $2.92.
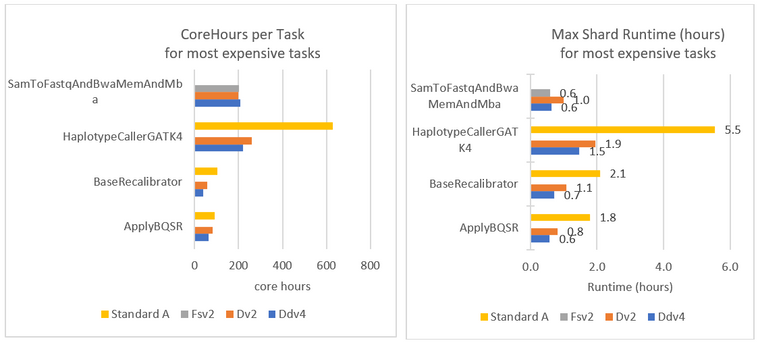
Figure 5 shows the performance in corehours consumed (A) and max shard runtime (B) for the four most expensive tasks in the pipeline. Figure 5A shows the sum of all corehours consumed across all VMs for a given task, while Figure 5B shows the maximum VM runtime of all VMs for that task. It is clear from both dimensions that the Dv2 and Ddv4 VM families are notably more performant than the Standard A series. While this is expected, the performance difference is so substantial that the longer runtimes are proportionally greater than the $/hour cost savings of the Standard A machines.
Figure 4
Figure 5
Figures 4 and 5 also breaks down the cost and performance for SamToFastqAndBwaAndMba (BWA) on the Fsv2 VM series. This CPU-bound task has the best cost/performance profile when running on the Compute Intensive Fsv2 Azure VM series, which is based on 1st and 2nd Generation Intel® Xeon® Scalable Processors, amongst the latest generation Intel CPUs available on Azure.
Conclusion
GATK Best Practices Pipeline for Germline Variant Calling is a critical genomics analytics workload and is broadly used in the fields of precision medicine and drug discovery.
Notably, running this pipeline on the Ddv4 Azure VM series with 2nd Generation Intel® Xeon® Scalable Processors results in both better performance and lower total cost when compared to running on the lowest priced VM ($/hour). Further cost savings can be achieved by specifically selecting the Fsv2 VM series for running the CPU-bound BWA-based tasks.
While we often assume trade-offs must be made between cost and time-to-solution, this work illustrates that when performance improvements are proportionally greater than cost savings (in terms of $/core-hour), one can have their cake and eat it.
Directly attributable to this work, Microsoft has contributed modifications to the Task Execution Service schema of the Global Alliance for Genomics and Health project to accept additional runtime attributes for each task. This will enable Cromwell on Azure to specify VM sizes for each task in a workflow and provides users the ability to achieve optimal performance and cost efficiency for their critical genomics workloads on Azure.
Configuration Details
Config: Intel Ddv4; Test By: Intel; CSP/Region: Azure uswest2; Virtual Machine Family: Ddv4; #vCPUs: {2, 4, 8, 16}; Number of Instances or VMs: 181; Iterations and result choice: Three; Median; CPU: 8272CL; Memory Capacity / Instance (GB): {8, 16, 32, 64}; Storage per instance (GB): {75, 150, 300, 600}; Network BW per instance (Mbps) (read/write): {1000, 2000, 4000, 8000}; Storage BW per VM (read/write) (Mbps): {120, 242, 485, 968}; OS: Linux; Workload and version: https://hub.docker.com/r/broadinstitute/genomes-in-the-cloud/; Libraries: GATK 4.0.10.1, GKL 0.8.6, Cromwell 52, Samtools 1.3.1; WL-specific details: Cromwell with Azure Batch; https://github.com/microsoft/gatk4-genome-processing-pipeline-azure
Config: Default; Test By: Intel; CSP/Region: Azure uswest2; Virtual Machine Family: A, Av2, Dv2, Dv3, Ls, Fsv2; #vCPUs: {1, 2, 4, 8, 16}; Number of Instances or VMs: 181; Iterations and result choice: Three; Median; CPU: E5-2660 (A); E5-2660, E5-2673 v4 (Av2); 8272CL, 8171M, E5-2673v4, E5-2673v3 (Dv2, Dv3); E5-2673 (Ls); 8168, 8272CL (Fsv2); Memory Capacity / Instance (GB): 2, 3.5, 7, 8, 32}; Storage per instance (GB): {10, 50, 100, 128, 135, 285, 678}; Network BW per instance (Mbps) (read/write): {250 - 8000}; Storage BW per instances (read/write) (Mbps): {20/10 – 750/375}; OS: Linux; Workload and version: https://hub.docker.com/r/broadinstitute/genomes-in-the-cloud/; Libraries: GATK 4.0.10.1, GKL 0.8.6, Cromwell 52, Samtools 1.3.1; WL-specific details: Cromwell with Azure Batch; https://github.com/microsoft/gatk4-genome-processing-pipeline-azure
Config: Dv2; Test By: Intel; CSP/Region: Azure uswest2; Virtual Machine Family: Dv2; #vCPUs: {1, 2, 4, 8, 16}; Number of VMs: 181; Iterations and result choice: Three; Median; CPU: 8272CL, 8171M, E5-2673v4, E5-2673v3; Memory Capacity / VM (GB): {3.5, 7, 14, 28, 56}; Storage per VM (GB): {50, 100, 200, 400, 800}; Network BW per VM (Mbps) (read/write): {750, 1500, 3000, 6000, 12000}; Storage BW per VM (read/write) (Mbps): {46/23, 93/46, 187/93, 375/187, 750/375}; OS: Linux; Workload and version: https://hub.docker.com/r/broadinstitute/genomes-in-the-cloud/; Libraries: GATK 4.0.10.1, GKL 0.8.6, Cromwell 52, Samtools 1.3.1; WL-specific details: Cromwell with Azure Batch; https://github.com/microsoft/gatk4-genome-processing-pipeline-azure
Posted at https://sl.advdat.com/3jF9zE7